Version 2024
This documentation is for PKanalix.
©Lixoft
PKanalix performs analysis on PK data set including:
- The non compartmental analysis (NCA) – computation of the NCA parameters using the calculation of the \(\lambda_z\) – slope of the terminal elimination phase.
- The bioequivalence analysis (BE) – comparison of the NCA parameters of several drug products (usually two, called ‘test’ and ‘reference’) using the average bioequivalence.
- The compartmental analysis (CA) – estimation of model parameters representing the PK as the dynamics in compartments for each individual. It does not include population analysis performed in Monolix.
What else?
- A clear user-interface with an intuitive workflow to efficiently run the NCA, BE and CA analysis.
- Easily accessible PK models library and auto-initialization method to improve the convergence of the optimization of CA parameters.
- Integrated bioequivalence module to simplify the analysis
- Automatically generated results and plots to give an immediate feedback.
- Interconnection with MonolixSuite applications to export projects to Monolix for the population analysis.
PKanalix tasks
Pkanalix uses the dataset format common for all MonolixSuite applications, see here for more details. It allows to move your project between applications, for instance export a CA project to Monolix and perform a population analysis with one “click” of a button.
Non Compartmental Analysis |
Bioequivalence |
Compartmental Analysis |
| This task consists in defining rules for the calculation of the \(\lambda_z\) (slope of the terminal elimination phase) to compute all the NCA parameters. This definition can be either global via general rules or manual on each individual – with the interactive plots the user selects or removes points in the \(\lambda_z\) calculation. |
The average NCA parameters obtained for different groups (e.g a test and a reference formulation) can be compared using the Bioequivalence task. Linear model definition contains one or several fixed effects selected in an integrated module. It allows to obtain a confidence interval compared to the predefined BE limits and automatically displayed in intuitive tables and plots. |
This task estimates parameters of a pharmacokinetic model for each individual. The model can be custom or based on one of the user-friendly libraries. Automatic initialization method improves the convergence of parameters for each individual. |
All the NCA, BE and/or CA outputs are automatically displayed in sortable tables in the Results tab. Moreover, they are exported in the result folder in a R-compatible format. Interactive plots give an immediate feedback and help to better interpret the results.
The usage of PKanalix is available not only via the user interface, but also via R with a dedicated R-package (detailed here). All the actions performed in the interface have their equivalent R-functions. It is particularly convenient for reproducibility purpose or batch jobs.
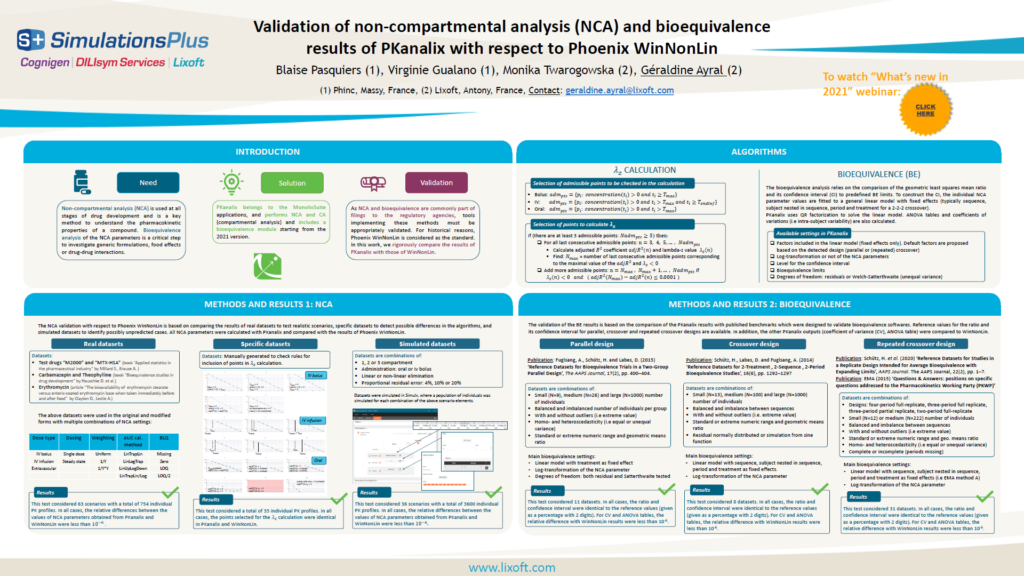
The results of the NCA calculations and bioequivalence calculations have been compared on an extensive number of datasets to the results of WinNonLin and to published results obtained with SAS. All results were identical. See the poster below for more details.