This page describes the settings of the bioequivalence calculations.
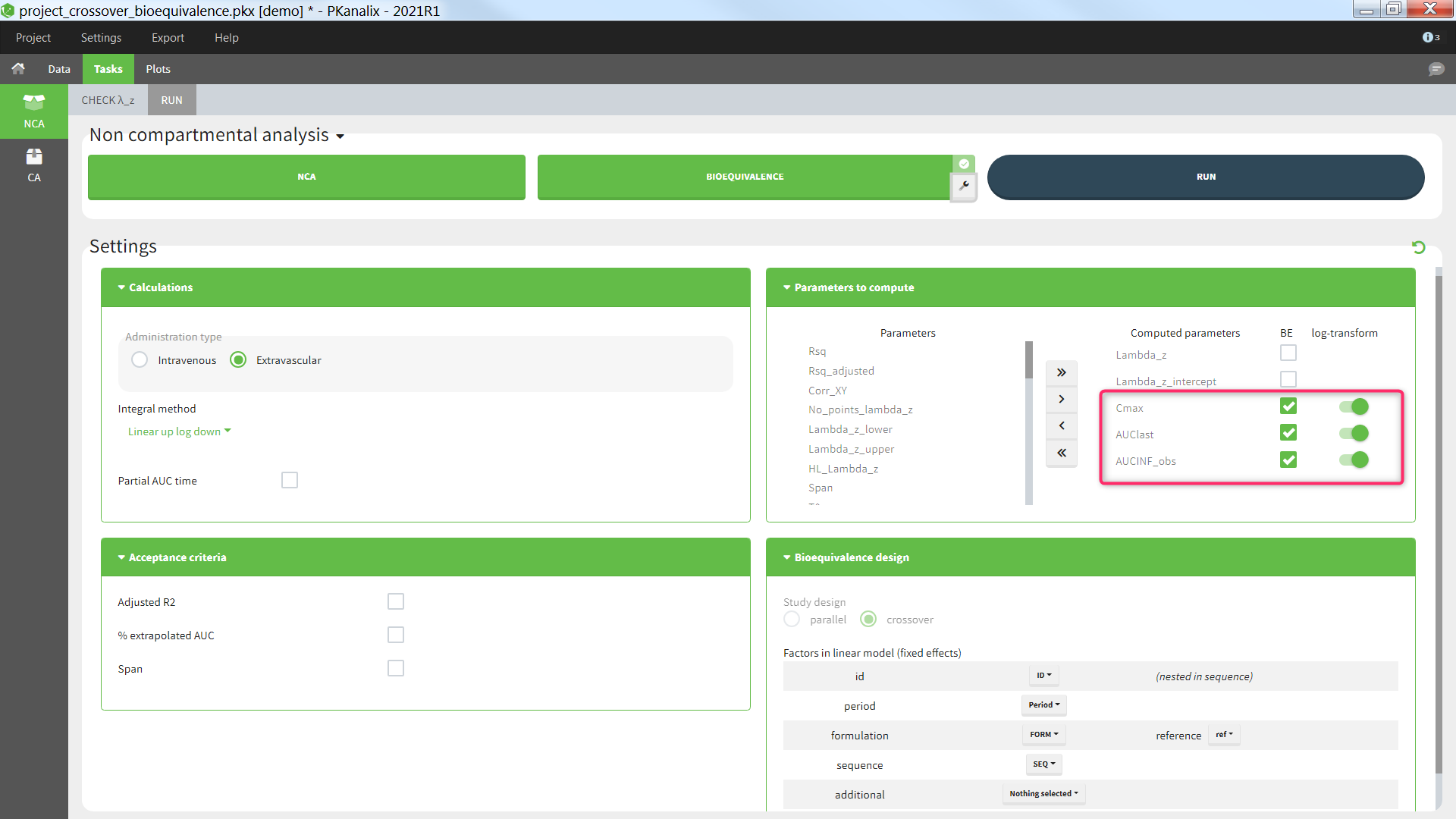
Parameters to compute
In the section “Parameters to compute”, the user can move the parameters from the “Parameters” column (list of all available parameters) to the “Computed parameters” column to select them for the NCA calculations. Among the parameters selected for the NCA calculations, the user can choose which parameters should be included in the bioequivalence analysis by ticking the box in the “BE” column. Regulatory guidelines usually recommend to perform the bioequivalence on the log-transformed NCA parameters. This can be set using the toggles in the “log-transform” column.
- BE: parameters on which to perform the bioequivalence analysis. Defaults: Cmax, AUClast, and AUCINF_obs if plasma single-dose, or AUCtau and Cmax if plasma steady-state, or AURC_last and Max_Rate if urine data.
- log-transform: whether the bioequivalence should be computed on the log-transformed NCA parameter (toggle on) or on the NCA parameter directly (toggle off). Default: log-transform toggle on, except for Tmax.
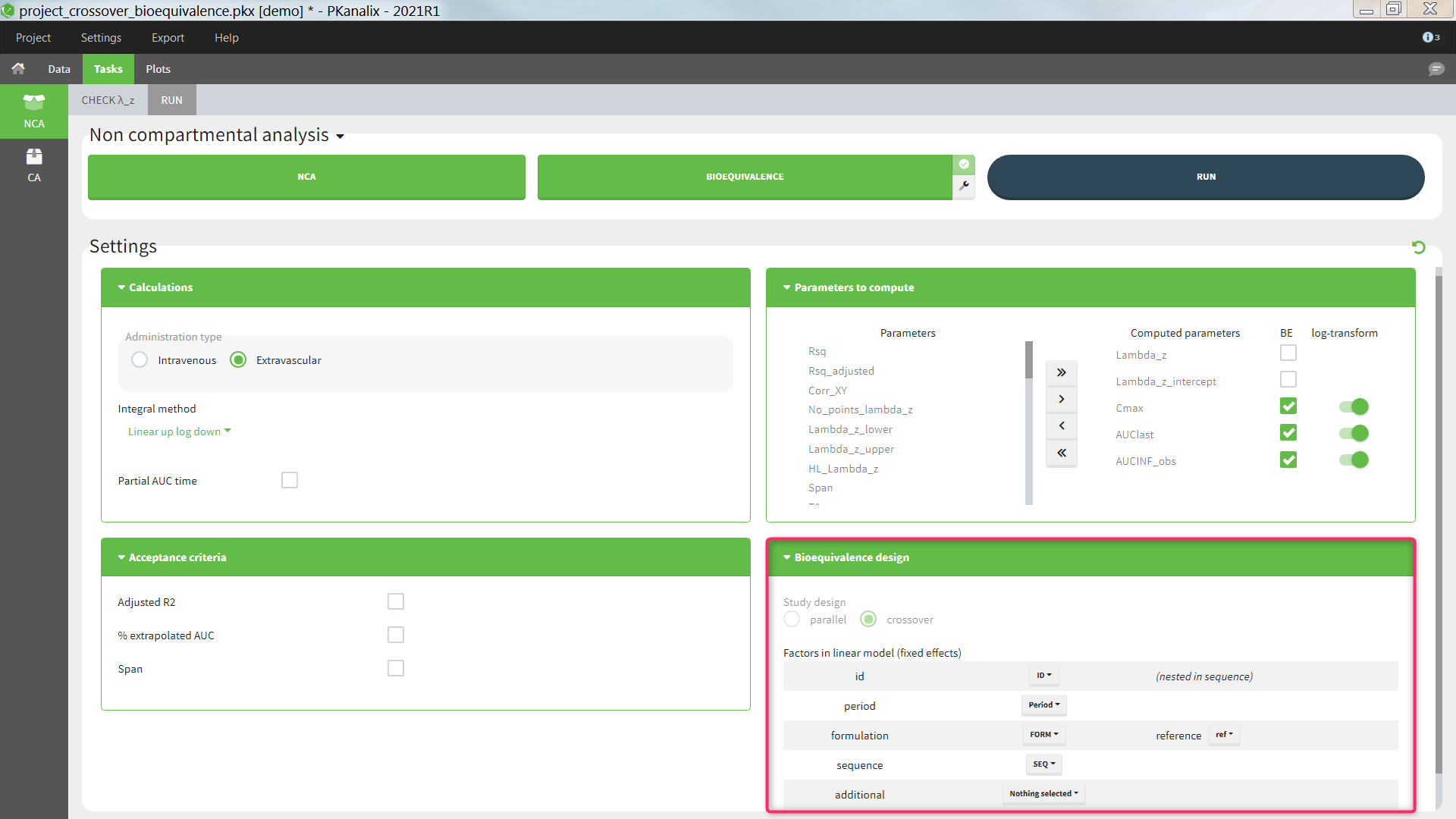
Bioequivalence design
The study design, parallel or crossover (repeated or not), is detected automatically based on the structure of the ID and OCCASION columns of the data set, see the Calculation rules for more details. The user can then select one or several fixed effect factors to be included in the linear model. By default, for a parallel design, only the formulation is included. For a crossover design, the factors ID, PERIOD, FORMULATION and SEQUENCE are included by default, if present in the data set. When both ID and SEQUENCE are given, ID is considered as nested in SEQUENCE. Additional factors can also be added, to be chosen among the (STRATIFICATION) CATEGORICAL and CONTINUOUS COVARIATES columns of the data set. All factors are considered as fixed effects and random effects are not available. Nesting of ID in SEQUENCE is considered automatically, but further nesting cannot be specified. Note that the nesting affects the calculation of the adjusted means but not of the bioequivalence ratio.
- Study design: parallel or crossover (repeated or not-repeated). Detected automatically based on the structure of the ID and OCCASION columns of the data set (see Calculation rules). Cannot be changed by the user.
- Factors in the linear model:
- id [if crossover]: data set column representing the ID of the individuals. Default: the column tagged as ID in the data set. This can be set to “none” if the ID should not be used as a factor in the linear model. When a “sequence” column is defined, “id” is considered as nested in “sequence”.
- period [if crossover]: data set column representing the different periods for each individual. Default: the first column tagged as OCCASION in the data set. This setting can be set to “none” if the period should not be used as a factor in the linear model.
- formulation: data set column representing the different groups for which the bioequivalence is calculated, typically the different formulations of the drug products. Default: the column with header ‘trt’, ‘treatment’, ‘form’, or ‘formulation’ (case insensitive) if it exists, otherwise first CATEGORICAL COVARIATE column. It is mandatory to include the formulation in the linear model.
- reference: among the categories of the categorical covariate selected as formulation, the one considered as the reference. The bioequivalence will be computed for all categories compared to the reference category. Default: ‘r’ or ‘ref’ or ‘reference’ (case insensitive), otherwise the category of the first occasion of the first indiv of the data set.
- sequence [if crossover]: data set column representing the different sequences. Default: the column with header ‘seq’, or ‘sequence’ (case insensitive) if it exist, otherwise “none”. This setting can be set to “none” if the sequence should not be used as a factor in the linear model.
- additional: data set columns tagged as CATEGORICAL or CONTINUOUS covariates, which have not already been selected above, can be added as additional factors in the linear model.
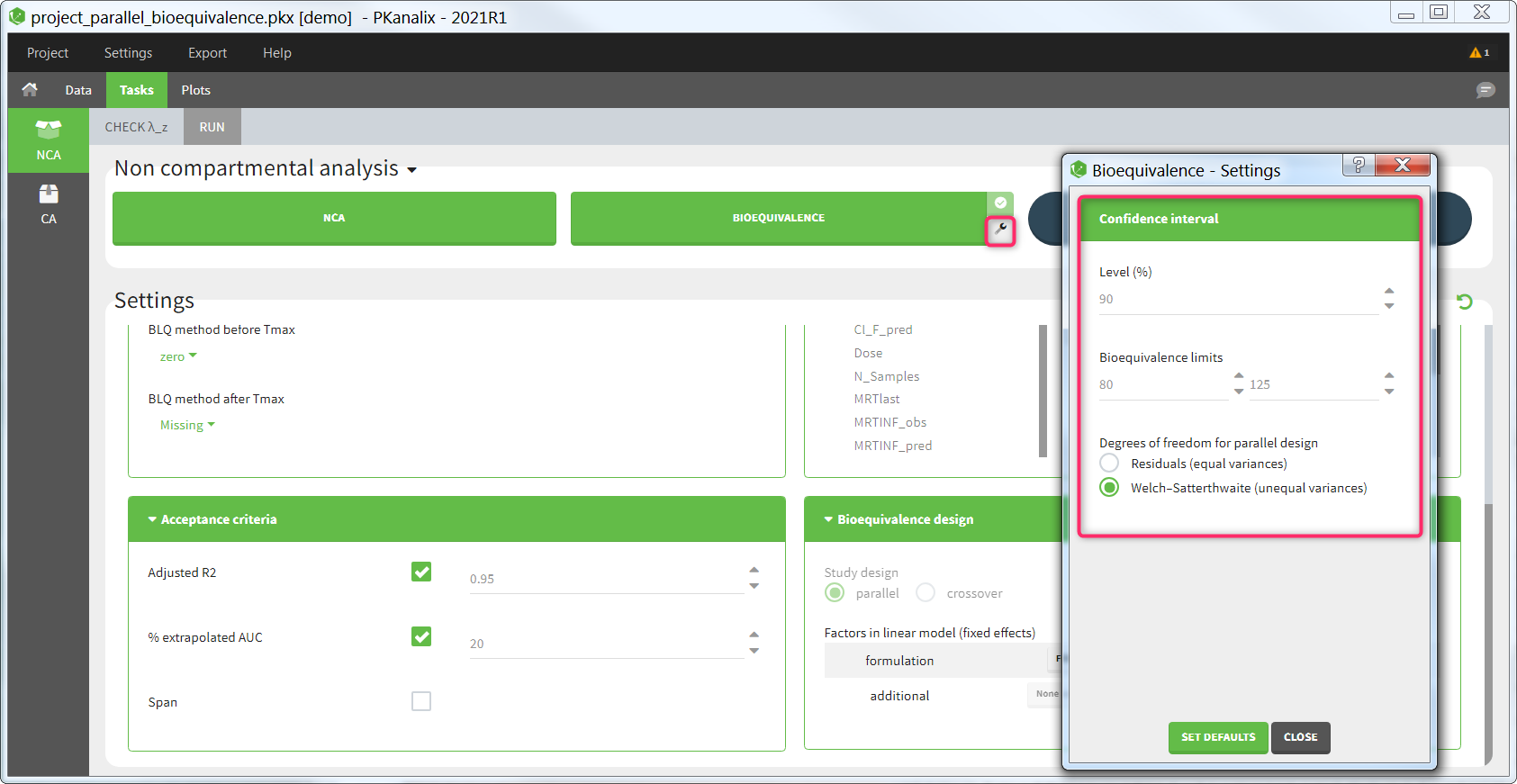
Confidence interval
Several settings are more specific to the calculation of the confidence interval. They can be changed in the pop-up settings window of the Bioequivalence task.
- Level: level of the confidence interval. Default: 90% confidence interval. This means that there is a 90% probability that the true ratio is included in the calculated confidence interval.
- Bioequivalence limits: limits within which the confidence interval should be in order to conclude for bioequivalence. Default: [80,125].
- Degrees of freedom [if parallel design]: method to calculate the degrees of freedom involved in the confidence interval calculations, either using the residuals (this assumes equal variance between the groups or using the Welch-Satterthwaite formula (this assumes possibly unequal variances). Default: Welch-Satterthwaite