PKanalix provides libraries of ready-to-use models for compartmental analysis. This simplifies the use of the software by decreasing the necessity of coding. You can pick models from the libraries, use them directly or edit. They are accessible in the Model sub-tab of the CA task by clicking the button “LOAD FROM LIBRARY“.
Libraries are organized into several categories:
- PK model library includes standard compartmental models. It is possible to select different administration routes (bolus, infusion, first-order absorption, zero-order absorption, with or without Tlag), different number of compartments (1, 2 or 3 compartments), and different types of eliminations (linear or Michaelis-Menten). The PK library models can be used with single or multiple doses data, and with two different types of administration in the same data set (oral and bolus for instance).
- PD model library includes direct response models such as Emax and Imax with various baseline models, and turnover response models. These models are PD models only and the drug concentration over time must be defined in the data set and passed as a regressor.
- PK/PD model library provides all standard combinations of pharmacokinetic and pharmacodynamic models.
- PK double absorption model library includes double absorption models. They take into account all the combinations of absorption types and delays for two absorptions. The absorptions can be specified as simultaneous or sequential, and with a pre-defined or independent order.
- Parent-Metabolite model library includes models describing parent drug and one metabolite, with or without the first pass effect, uni and bidirectional transformation and up to three compartments for parent and metabolite.
- Target-mediated drug disposition (TMDD) model library – includes a large number of TMDD models corresponding to different approximations, different administration routes, different parametrizations, and different outputs.
- Tumor growth inhibition (TGI) library – includes a wide range of models for tumour growth (TG) and tumour growth inhibition (TGI) that are available in the literature. Models correspond to different hypotheses on the tumor or treatment dynamics.
In each library, available models are displayed as a list. With the top filtering panel you can easily selected a model based on its characteristics such as administration type, number of compartments or elimination mechanism.
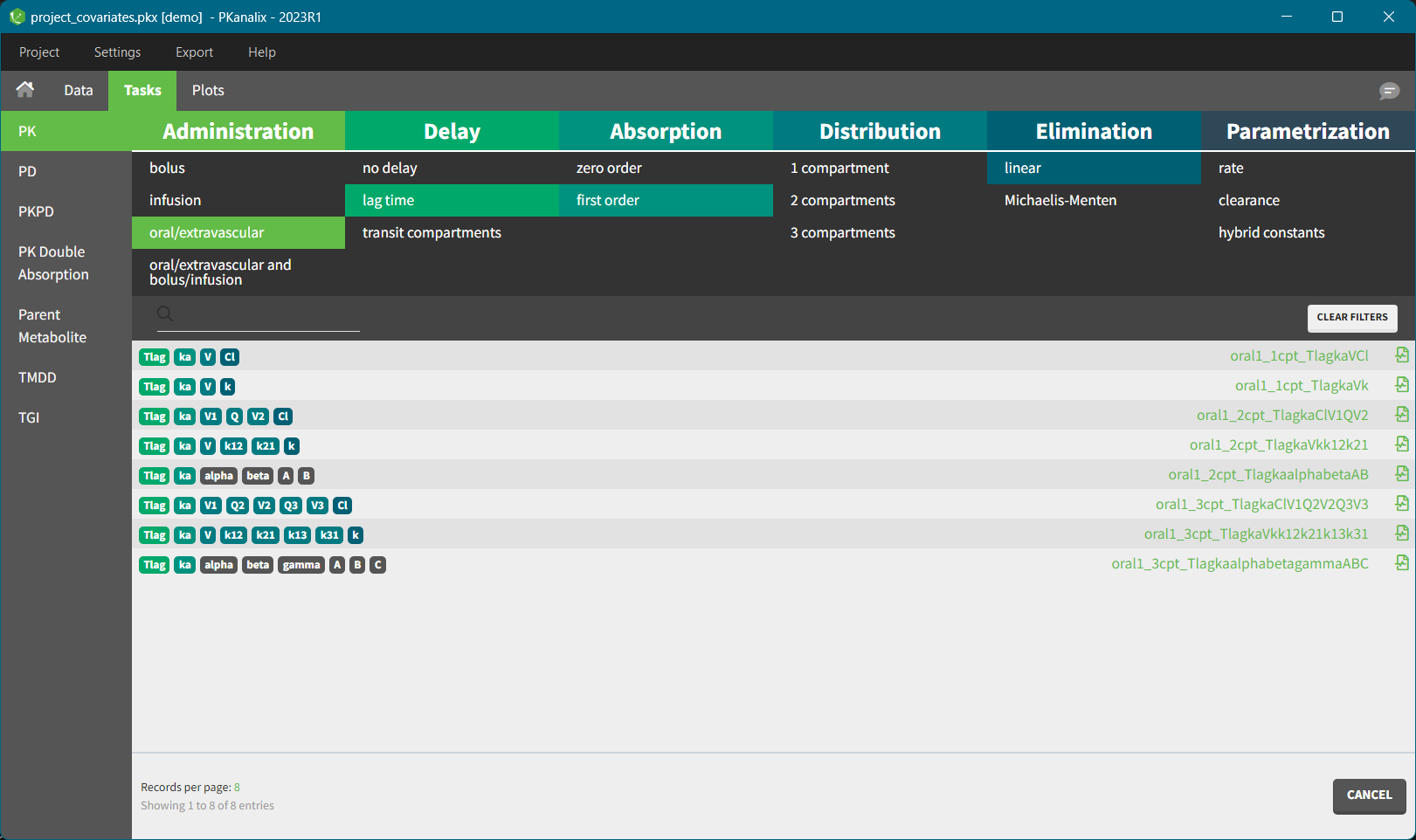
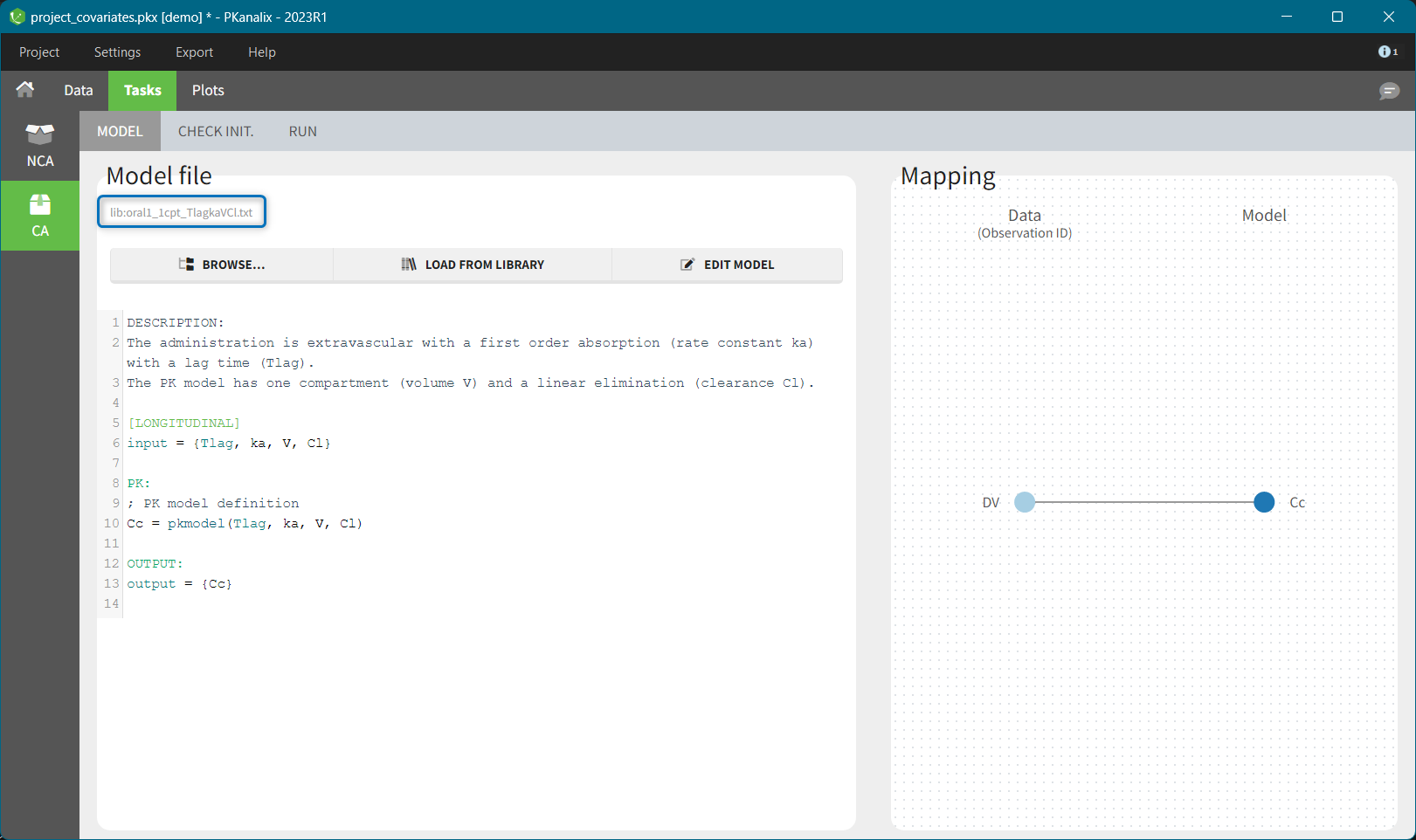
To select a model click on its corresponding line – when all filters are used, only one model is shown. The model will be automatically loaded and displayed in the Model tab. Model filename is displayed on top of the “Model file” section, and has a prefix “lib” to distinguish it from custom models.
Next section: Custom model