We provide an Excel macro that can be used to adapt the formatting of your dataset for use in PKanalix:
Download the Excel macro to adapt data formatting for PKanalix (version March 21st 2022)
In case of download issue: the download may be blocked by your browser which identifies this type of file as potential threat. Right-click the link and choose “Save link as”. After validating, if the browser displays “the file can’t be downloaded securely”, click on the arrow to display more options and choose “Keep”. Once the file is downloaded, right-click the file, open its Properties and select the Unblock checkbox at the bottom of the General tab, and select OK.
The macro is not compatible with Excel on Mac.
The macro takes as input the original dataset, in Excel or text format, adds the dosing information separately and produces an adapted dataset with the doses in a new column. The macro can also translate information on missing observations, censored observations, urine volume, and occasions into the PKanalix-compatible format. The adapted dataset is saved as an Excel file and, if selected, as a CSV file for direct use in PKanalix.
More details of the actions performed by the macro are described below.
Steps to follow to adapt the data format:
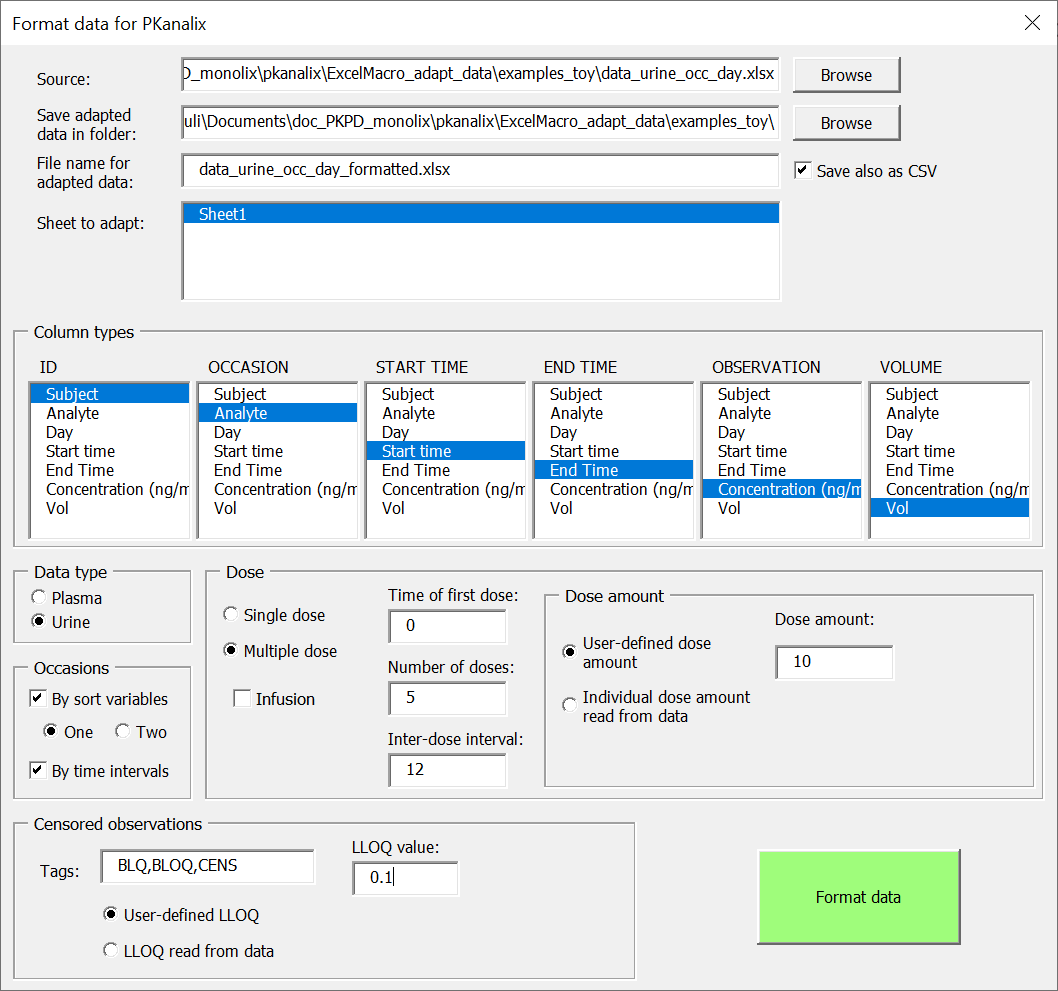
- Click on the button “Select file to adapt” and choose the dataset file to open in Excel
- Fill the form to give information on the dataset:– In case of several sheets, select the name of the sheet to adapt
- Select the columns containing ID, TIME and OBSERVATION information
- If individual concentrations should be split in different profiles by occasions, check the option “Occasions: by sort variables”, select one or two levels, and select the column(s) containing the OCCASION information
- If individual concentrations should be split by intervals of successive times, check the option “Occasion: by time intervals”
- In case of urine data, change the corresponding radio button and select the columns containing each collection interval START TIME and END TIME, and the column containing VOLUME information.
- Give treatment information: single/multiple dose, time of first dose, dose amount (individual dose amounts can be read from a column of the dataset), number of doses, inter-dose interval, and infusion duration in case of IV infusion.
- In case of censored observations, indicate the tag(s) representing censored observations in the dataset if it is different than the default tags (several tags can be entered, separated by commas without spaces), and give the value of LLOQ or the header of a column containing LLOQ values.
- Click on “Format data” and wait for the result.
Result:
- The adapted dataset is saved in a new .xlsx file with a name derived from the original name or user-specified.
- If requested in the form, the adapted dataset is also saved in a new .CSV file. If the initial file has several sheets, the sheet name is added to the CSV file name.
- If the file already exists, a new sheet is added to the file, unless a sheet with the same name already exists.
- All changes from the original dataset are highlighted with colors:
— blue = dosing time or amount,
— gray = missing observation,
— orange = censored observations,
— green = urine volume at dosing time,
— yellow = new occasions as integers.
Actions performed by the macro:
- A column AMT is added, containing the same dosing regimen for all subjects or subject-occasions (with an option to read individual dose amounts from a column of the dataset). The column is colored in blue, as well as dosing times. Note that the dosing times are correct in the column tagged as TIME in the form, but other time columns will not be consistent.
- In case of urine data, the volume is set to 0 at dosing times (colored in green) to avoid format errors, and both start and end times of the interval are set to the dosing time. The macro checks that time intervals are continuous (no gaps allowed). Only the column END TIME should be used as TIME in PKanalix.
- In case of censored observations, a column CENS is added and colored in orange, containing 0 for non-censored observations and 1 for censored observations. The LLOQ is written instead of the censoring tag in the column of observations and colored as well.
- Missing observations (all values that are not numbers) are replaced by “.” and colored in gray.
- In case of occasions, for each column tagged as containing occasions, a new column with the same header concatenated with “_OCC” is added with occasion numbers and colored in yellow. If the column tagged already contained numbers, this just duplicates the column, but the initial column can then be used as categorical covariate.
- In case of occasions by time intervals, two identical columns named Interval_OCC and Interval_CATCOV are created next to the column TIME, with indexes of intervals of successive times for each individual.
- The first lines that do not contain data are concatenated into a single header row.
- Rows that do not contain data are deleted.
- Formatting is adapted for correct exporting to CSV format.
Examples
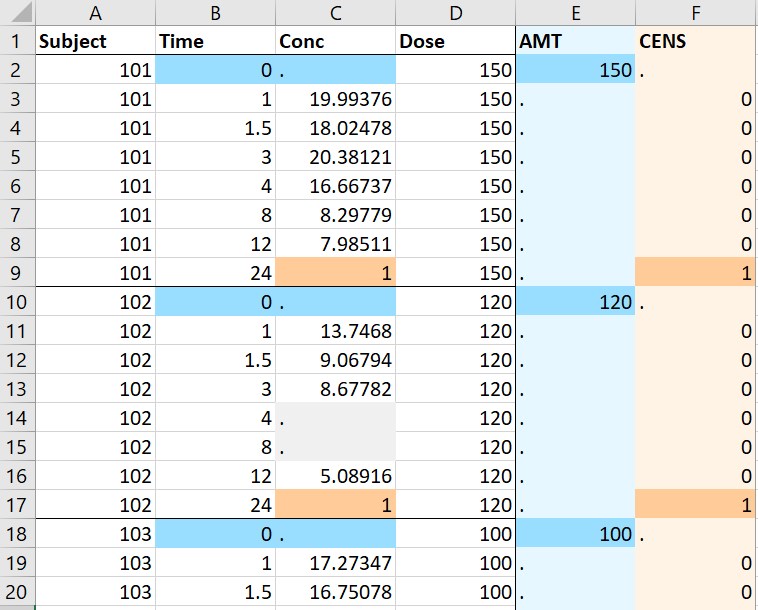
- Single doses: the original data on the left contains concentration data. All individuals have received single doses at time 0, with individual dose amounts in the column Dose. The macro is used to add the dosing times. In the form, the columns Subject, Time, Conc are tagged as ID, TIME and OBSERVATION respectively. The column Dose is indicated in the form as containing the dose amounts, and the single dosing time is also specified in the form. In addition to adding the dosing times and the corresponding amounts in a new column AMT, the macro replaces the missing observations (any value in Conc that is not a number) by . compatible with PKanalix. The macro form can also be used to specify that <BLQ> is a flag for censored observations rather than missing ones, with a LLOQ of 1. In that case the macro adds a new column CENS and replaces <BLQ> by the LLOQ. Finally, the second line containing units is removed from the adapted dataset, because it can not be read by PKanalix.
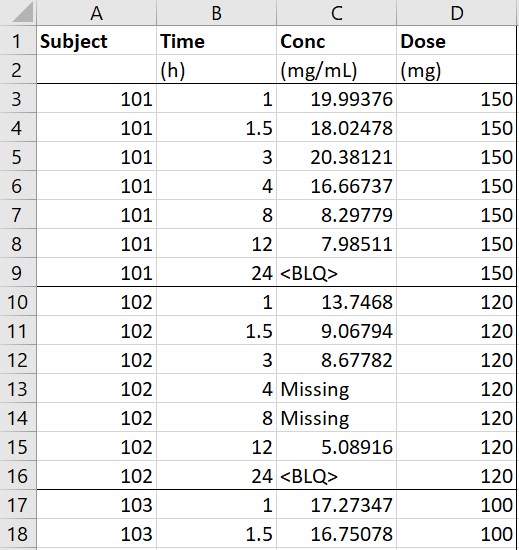
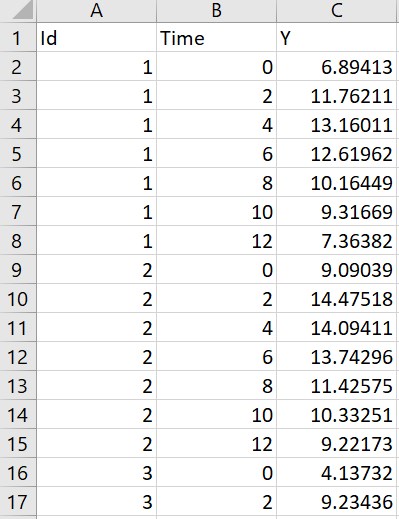
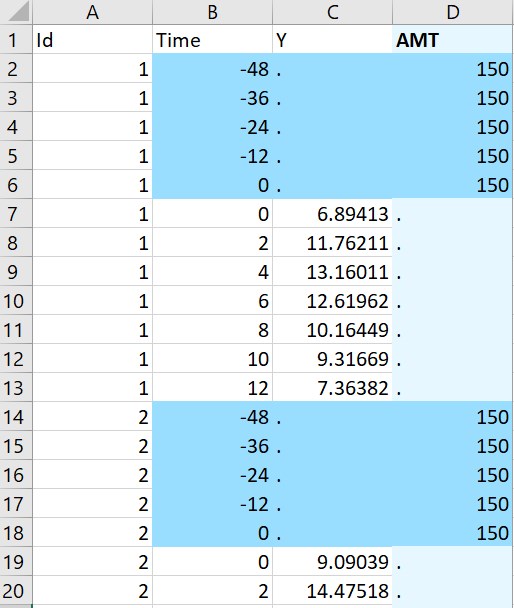
- Multiple doses: the original data on the left contains only ids, measurement times and concentrations. The macro is used to add doses. In the macro form, the columns Id, Time and Y are tagged as ID, TIME and OBSERVATION. A multiple dose regimen is specified with 5 doses starting at time -48 with interdose interval of 12 and level of 150. Running the macro with these settings adds a column AMT with 5 doses per individual.
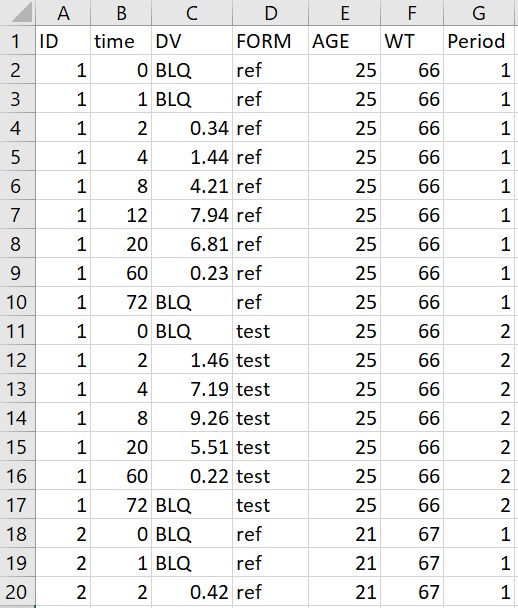
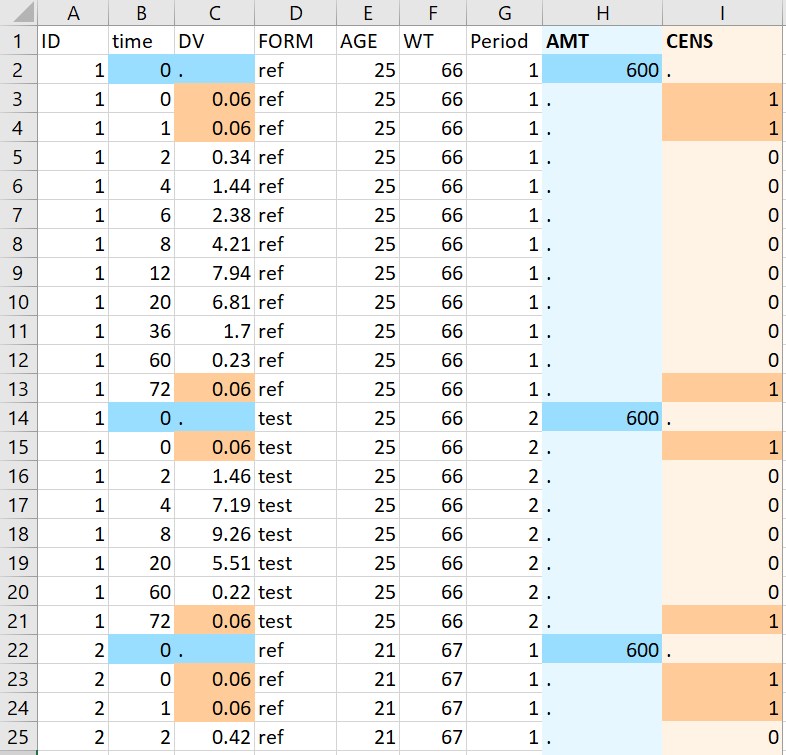
- Censored observations: on the example below (original data on the left, adapted data on the right), the column Period already contains occasions as integers, corresponding to the different values of FORM. Censored observations are flagged with “BLQ” in the column DV. The macro is used with the following settings: ID, time, DV and Period are tagged respectively as ID, TIME, OBSERVATION and OCCASION. A single dose is specified with value 600 at time 0, and the specified LLOQ value is 0.06. Running the macro adds two new columns AMT and CENS containing the doses and the censored observations flags. “BLQ” is replaced by 0.06 in the column DV. The column Period is left untouched since it is already compatible with PKanalix.
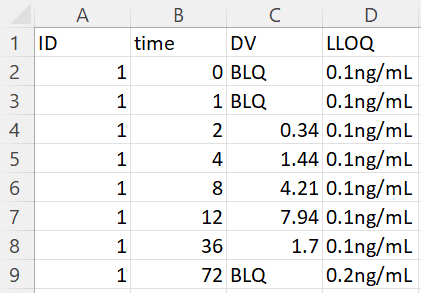
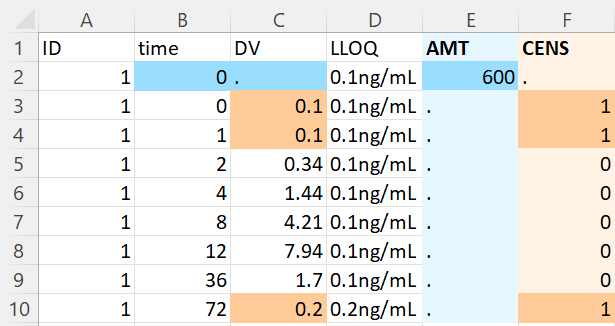
- Censored observations with LLOQ read from data: on the next example, the LLOQ value is not given manually in the form, but the option “read LLOQ from data” is selected. On each line, the line in the column LLOQ (selected in the form) is used as LLOQ value if the value on the line is censored. Notice that the value can be extracted from a string: all non-numeric characters are removed except “.”.
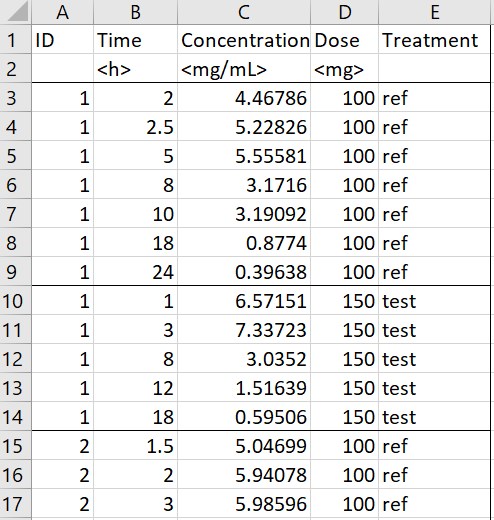
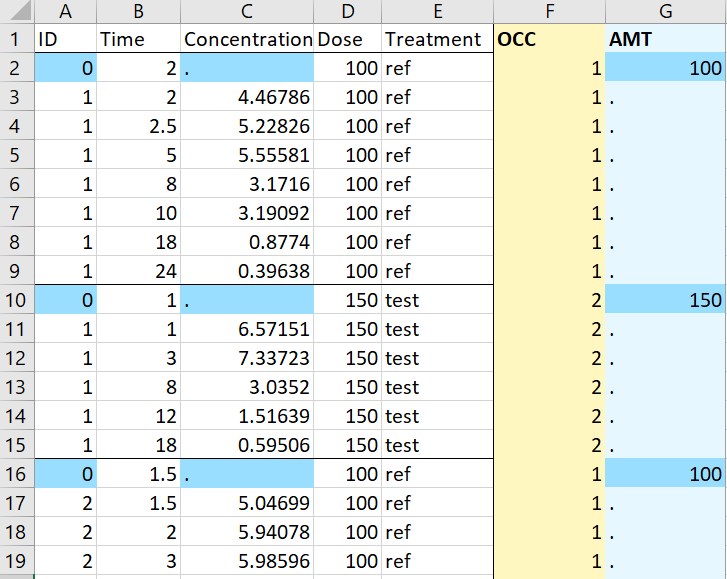
- Occasions by sort variable: the original data on the left contains a sort variable (or time-varying covariate) “Treatment”, which should be defined in different occasions for PKanalix. Running the macro with “Treatment” tagged as “Occasion” creates a new column OCC in the adapted dataset with different integer occasions corresponding to the different values of “Treatment”. Moreover, the dose level is read for each subject-occasion from the column “Dose”. Here a single dosing time at 0 has been specified in the macro form. Finally, the second line containing units is removed from the adapted dataset, because it can not be read by PKanalix.
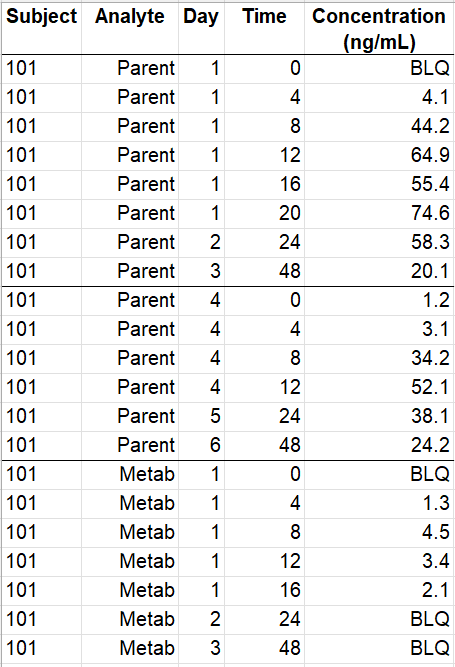
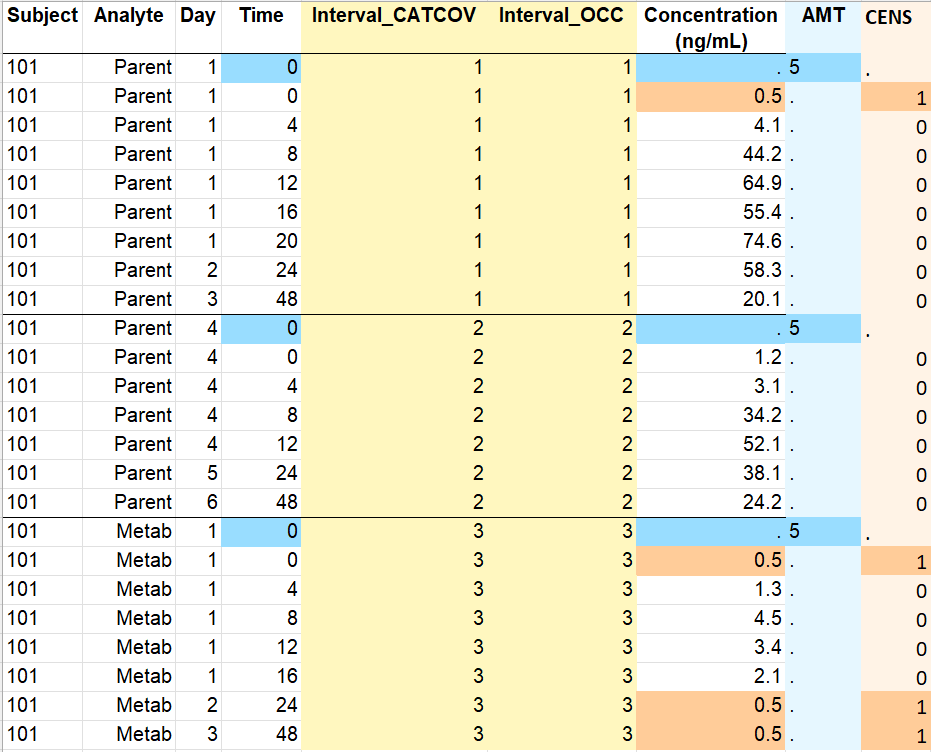
- Occasions by time intervals: the original data on the left contains concentration profiles measured on different days with the time after the last dose in hours. Two doses have been given on days 1 and 4, and the drug concentration is measured on days 1 to 6. Selecting the option “Occasions as time intervals” in the form creates the columns Interval_CATCOV and Interval_OCC that distinguish the concentration profiles with the indexes of intervals of successive times. Lines are assigned to a time interval by order, with a change of interval when the id changes or when the time does not increase between a line and the next. Here it is not necessary to select the column Analyte as OCCASION (or sort variable): since concentration profiles from parent and metabolite are separated in order with different intervals of successive times, they are also distinguished with the Interval columns. The form should also by used to specify the doses (single dose at time 0 with amount 5) and the LLOQ (0.5) with BLQ as censoring tag.
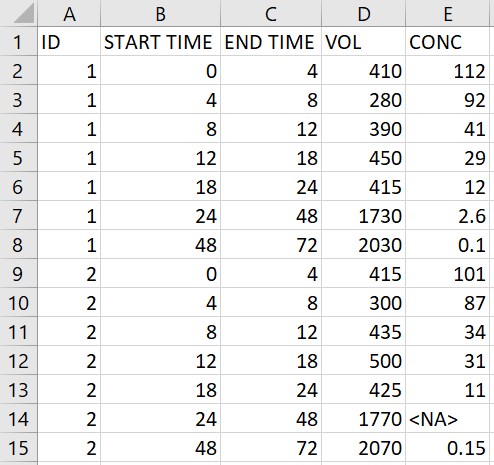
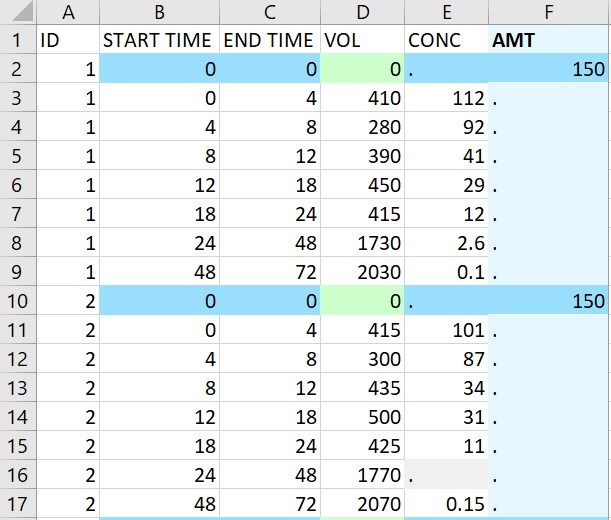
- Urine data: the original data on the left contains urine concentration measurements in the column CONC, with the urine volume in VOL collected during the interval defined by START TIME and END TIME. Using the macro to add a single dose of 150 at time 0 to all individuals produces the data on the right. Since VOL is tagged as urine volume in the macro, the value 0 for VOL is added at dosing times, and the macro checks that the intervals are continuous. The missing concentration for the interval between times 24 and 48 for id 2 is replaced by a . compatible with PKanalix.